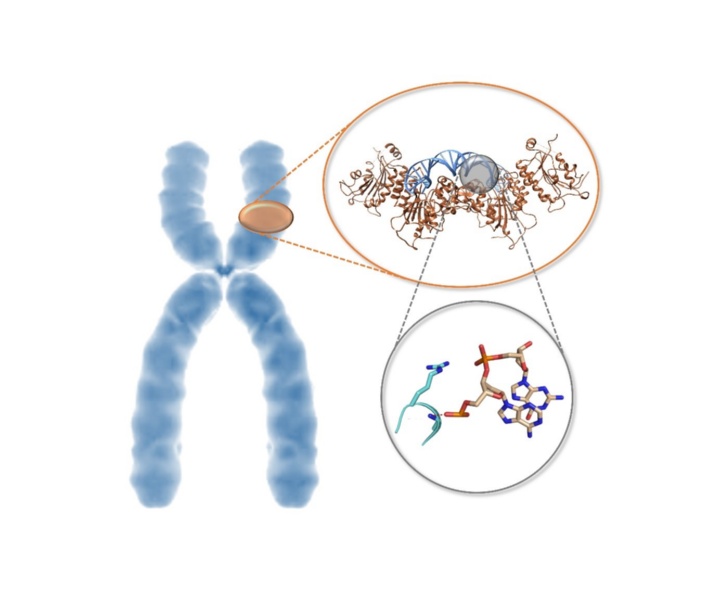
The ICF syndrome is a genetic developmental disorder that, among other things, leads to an immune deficiency and therefore increases the susceptibility of those affected to other diseases. Pioneering genetic observations around 20 years ago showed that the key to this disease lies in inadequate activity of the DNMT3B enzyme. An international research team led by Prof. Albert Jeltsch from the Institute of Biochemistry and Technical Biochemistry (IBTB) at the University of Stuttgart has now been able to explain these relationships from a biochemical-molecular point of view.
In the course of the evolution of higher organisms such as mammals, gene duplication often occurs. The resulting "twin genes" have a very similar genetic makeup, but are independent of each other and can therefore specialize in certain tasks. An example of this is DNA methylation, a chemical change in the basic building blocks of the genetic material of a cell, which is caused by the transfer of methyl groups by enzymes (DNA methyl transferases, DNMT) to certain locations in the DNA. The DNA-methyltransferases DNMT3A and DNMT3B are two forms of these enzymes in human cells that have arisen from gene duplication.
DNMT3B activity too low
Over 20 years ago, it was discovered that DNMT3B is essential for the DNA methylation of certain regions in the human genome and that insufficient activity of DNMT3B leads to the so-called ICF syndrome. However, why DNMT3B is specifically required for this task and why, for example, the twin DNMT3A cannot take over this task has so far remained unknown.
In cooperation with groups from the University of California and the University of North Carolina at Chapel Hill, the team led by Prof. Albert Jeltsch from the University of Stuttgart has now been able to solve the structure of DNMT3B in complex with other DNA sequences and measure the turnover rate of DNA methylation by DNMT3B and DNMT3A on thousands of DNA sequences. It was shown that DNMT3B is particularly active on its target sequences in the human genome due to a special protein loop, while DNMT3A can only work poorly on these sequences.
Role of DNMT3A
On the other hand, mutations in the same protein loop in DNMT3A also affect the properties of this enzyme, which leads to an increased susceptibility to tumors. “It is exciting to understand how complex functions develop on a molecular level during evolution. If one understands the role of DNMT3A better, one could in the future, for example, use an enzyme inhibitor in cancer therapy that curbs tumor growth” Jeltsch hopes.
Expert Contact:
Prof. Dr. Albert Jeltsch, University of Stuttgart, Institute of Biochemistry and Technical Biochemistry (IBTB), Phone +49 711 685 64390,
E-Mail
Originalpublikation:
Linfeng Gao et.al.: Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms, Nature Communications vom 3.7.2020, DOI 10.1038/s41467-020-17109-4, https://rdcu.be/b5o56