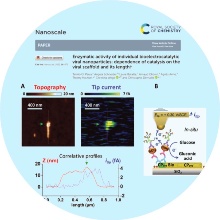
Die Forschungseinheit Molekulare und Synthetische Pflanzenvirologie beschäftigt sich mit Pflanzenvirus-Bausteinen für "Smart Materials" und Bionanotechnik in verschiedenen Anwendungsfeldern, molekularen Wechselwirkungen zwischen Pflanzen und Viren in Einzel- und Mischinfektionen sowie phytoviralem Engineering.
Neues aus der Pflanzenvirologie
Kontakt

Christina Wege
Prof. Dr. (apl.)Leitung Forschungseinheit Molekulare und Synthetische Pflanzenvirologie
- Profil-Seite
- +49 711 685 65073
- E-Mail schreiben
- Koordinatorin Projekthaus NanoBioMater & Fachstudienberatung

Tatjana Kleinow
PD Dr.Privatdozentin
[Foto: T. Kleinow]

Holger Jeske
Prof. (a.D.) Dr.vormals Leitung Abteilung Molekularbiologie und Virologie der Pflanzen; verstorben April 2022